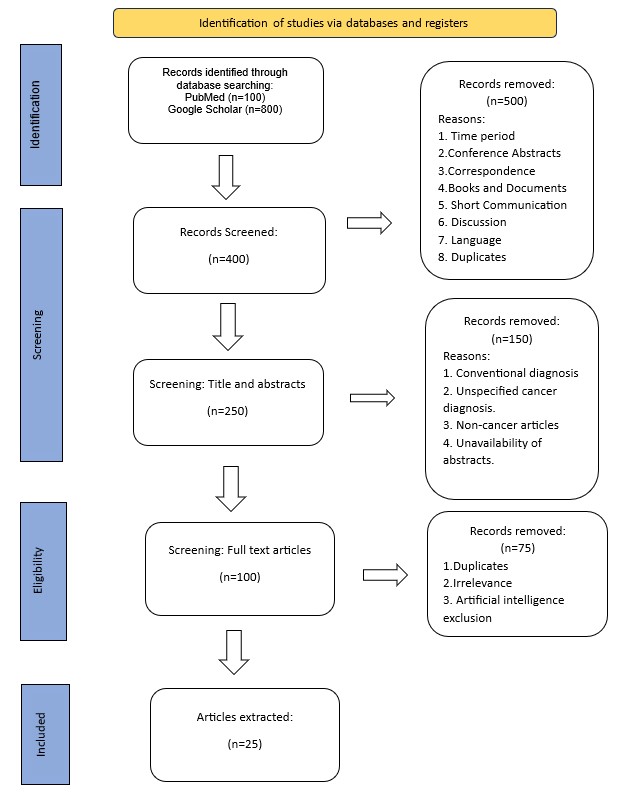
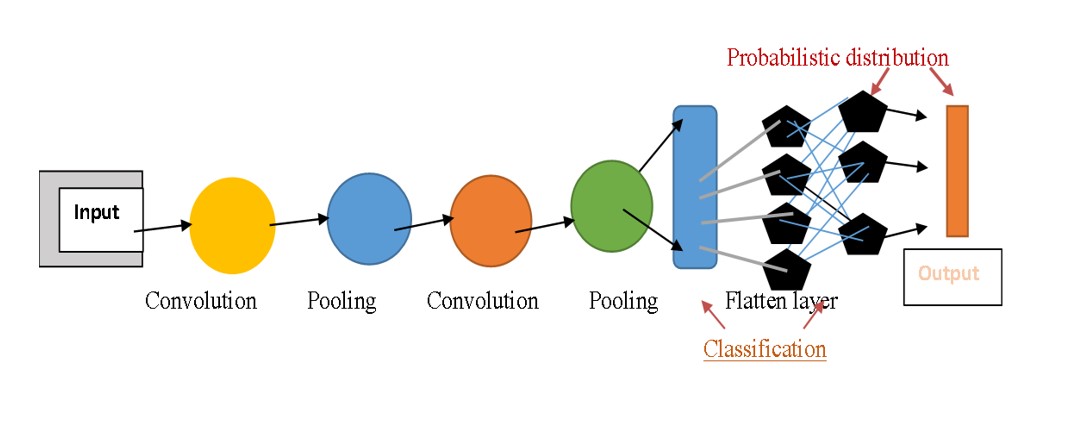
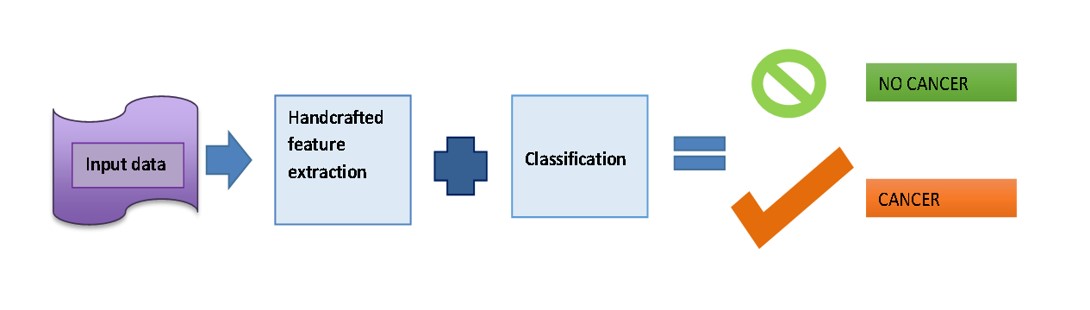
on its own by using artificial neural networks that mimic the structures and functions of the human brain. Deep Learning (DL) use deep neural networks (DNNs), which are inspired by the neural architecture of the brain, to build intricate models with several invisible layers that evaluate diverse varieties of input and predict outcomes. The machine receives raw data from DL algorithms so that it may train itself to automatically identify the top deep features for the job. This capability most likely explains why DL algorithms have continuously improved at performing many popular AI tasks, including speech recognition, identification of images, pattern recognition, and natural language processing. As a result, DL is used extensively in AI research in the realm of cancer. Oncologists have a keen interest in the application of AI to diagnose malignancies accurately using radiological imaging and pathological slides, forecast patient results, and make the best possible treatment decisions [1] (Figure 1).
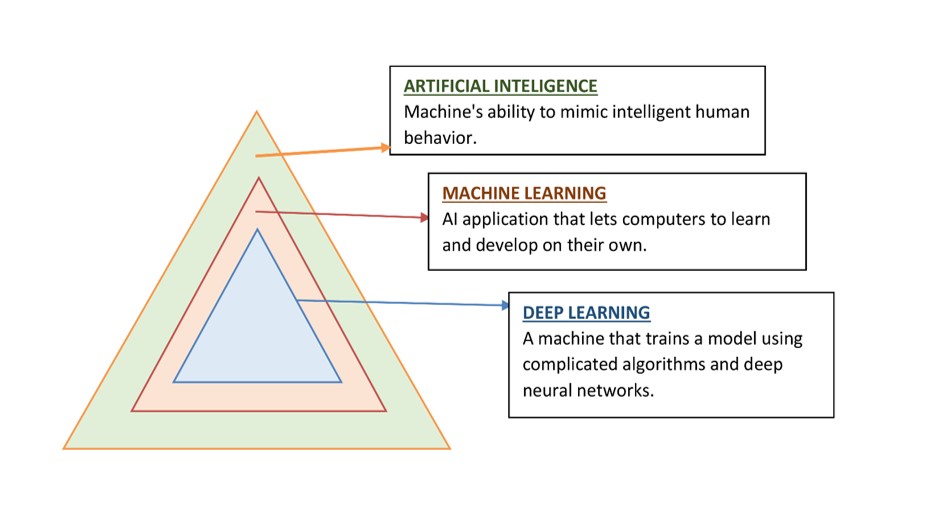
Figure 1: Artificial Intelligence Algorithms
Cancer ranks as the primary killer in both developed and developing countries and the death toll due to oncological diseases is expected to rise to 13.1 million by 2023. Delay in its detection often leads to poor prognosis and lowers the chances of survival. However, early detection of cancers can diminish its mortality rates [2]. Diagnosis of cancer can be divided into three categories: predicting the risk of occurrence of the disease, prediction of its recurrence and finally predicting the chances of survival of the cancer patients. Detection of cancers is most commonly done via medical imaging and histological methods. Different scanning techniques can be used to screen for oncology related diseases for example a computed tomography (CT) is used to detect the tumor shape and size. Similarly, nuclear scans such as gallium scans, positron emission tomography (PET scan), bone scans, multigated acquisition scans (MUGA) and thyroid scans help determine cancer metastasis. MRI is a commonly used tool to detect the occurrence and spread of the tumor. Mammograms and ultrasounds are widely used by specialists for screening breast cancers [3]. Interventional procedures including high definition white light endoscopy, traditional white light endoscopy, colonoscopy, endocytoscopy, confocal and endomicroscopy are used to detect polyps, lesions and to perform biopsies for confirmation of the type of cancer [4].
A main site of origin cannot be identified in metastatic cancer even after a routine diagnostic workup, which accounts for around 3-5% of all malignant illnesses. This condition is known as cancer of unknown primary (CUP). CUP patients suffer from substantial disadvantages and the most don't survive well since understanding a patient's original malignancy is still essential to their therapy, hence it is crucial to create reliable and approachable diagnostic techniques for determining the cancer's primary tissue of origin [5].
Methods
The following methodology (Figure 2) was adopted for the review.